This article on running Apache Spark on Kubernetes first appeared on Medium. This blog has been republished with the author’s credit and consent.
During my years working on Apache Spark applications, I have always been switching my environment between development and production. I would use an IDE such as Visual Studio Code to write the Scala or PySpark code, test it locally against a small piece of the data, submit the Spark job to Hadoop YARN to run in production, and hopefully, it just works on real big data. I would spend a lot of time dealing with Spark dependencies for my local and production environment and make sure they are in sync. This workflow becomes more painful for machine learning applications, where a combination of PySpark, Python libraries, and Jupyter Notebook environment is required. Even for enterprises with many engineers, it is still challenging to set up and maintain environments for Spark application development, data exploration, and running in production.
Enter Spark on Kubernetes. With Spark on Kubernetes, and ideally a fast object storage like FlashBlade® with S3, we can use a single environment to run all these different Spark tasks, easily.
- A Jupyter notebook backed by multiple Spark pods for quick prototyping and data exploration for small and big data.
- Declare a Spark application in a yaml file and submit it to run in production.
- Apache Airflow to orchestrate and schedule pipelines with multiple jobs.
This new workflow is much more pleasant compared to the previous one. All my tasks leverage the same Kubernetes environment. Dependencies are managed in container images so that they are consistent across development and production. Performance issues can be detected in the development phase because testing at scale becomes easier. And most importantly, there is no Hadoop cluster to manage anymore.
I explained how to set up Spark to run on Kubernetes and access S3 in my previous blog post. This time, I will describe my new workflow to run Spark on Kubernetes for development, data exploration, and production.
Supercharging Jupyter Notebook with PySpark on Kubernetes
Jupyter Notebook is handy for quick prototyping and data exploration because developers and data scientists can start coding right away on its web-based interactive development environment. However, since it runs in a single Python kernel, it could be slow processing big data. On the other hand, PySpark allows us to write Spark code in Python and run in a Spark cluster, but its integration with Jupyter was not there until the recent Spark 3.1 release, which allows Spark jobs to run natively in a Kubernetes cluster. This makes it possible to process big data from a Jupyter notebook.
With a couple of lines of code configuration, we can now write PySpark code in a Jupyter notebook and submit the code to run as a Spark job in a Kubernetes cluster.

Launch a Spark on Kubernetes session from Jupyter.
In the example notebook below, my PySpark code reads 112M records from a CSV file stored in FlashBlade S3 and then performs some feature engineering tasks. Due to the huge number of records, if running on a single process, this could be very slow.

Read 112M records from S3 into PySpark.
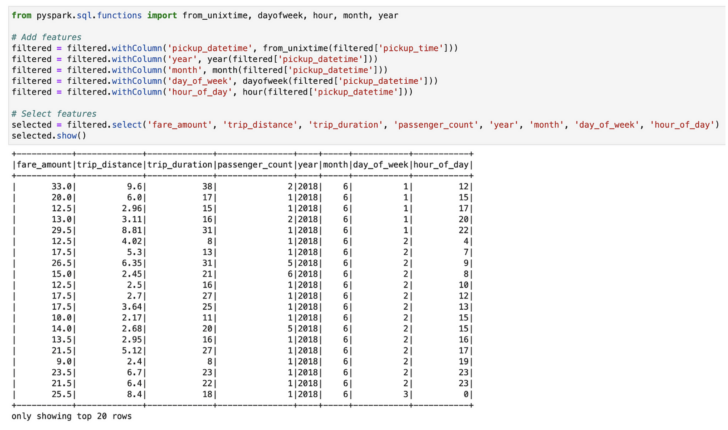
Feature engineering in PySpark.
However, in this case, on the backend, the heavy processing is handled by a Spark job running in Kubernetes. Below are the Spark pods in Kubernetes launched by the notebook.

Spark pods in Kubernetes launched by Jupyter notebook.
Because the backend is a fully distributed Spark job, it is fast. We can now process and explore a huge number of records in a Jupyter notebook from the browser.
Streamline Spark Submission in Production
The next step in the workflow is to submit the Spark code to production. There were discussions on whether or not a notebook should be treated as production code. Some companies such as Netflix have been doing this, but I think most are still not there yet. In my workflow, I would copy and refine the notebook code to a Python file, put it in S3, declare a PySpark job in a yaml file, and submit it to Kubernetes using the Spark on k8s operator. You can find the details in my previous blog post.
There are two approaches to submit a Spark job to Kubernetes in Spark 3.x:
- Using the traditional spark-submit script
- Using the Spark on k8s operator
I choose to use the Spark on k8s operator. Because it is native to Kubernetes, it therefore can be submitted from anywhere a Kubernetes client is available. With this approach, submitting a Spark job is a standard Kubernetes command: kubectl apply -f nyc-taxi.yaml. This helps streamline Spark submission. It is also more flexible because a Spark client is not required on the node.
You may have noticed that this is different from how I launched an ApacheSpark on Kubernetes session from Jupyter in the section above, where the traditional spark-submit is used. This is true because I wanted the Spark driver to run inside the Jupyter kernel for interactive development. Whereas in production, we want reproducibility, flexibility, and portability.
How to Optimized Deep Learning Workflows with S3
Orchestrate and Schedule Pipelines
While the Spark on k8s operator is great to submit a single Spark job to run on Kubernetes, oftentimes, we want to chain multiple Spark and other types of jobs into a pipeline and schedule the pipeline to run periodically. Apache Airflow is a popular solution for this.
Apache Airflow is an open source platform to programmatically author, schedule, and monitor workflows. It can run on Kubernetes. It also integrates well with Kubernetes. I will skip the details of how to run Airflow on Kubernetes, and from Airflow how to orchestrate Spark jobs to run on Kubernetes. For now, let’s focus on the behaviours and value it brings.
In the example below, I define a simple pipeline (called DAG in Airflow) with two tasks that execute sequentially. The first task submits a Spark job called nyc-taxi to Kubernetes using the Apache Spark on k8s operator; the second checks the final state of the Spark job that was submitted in the first state. I have also set the DAG to run daily.
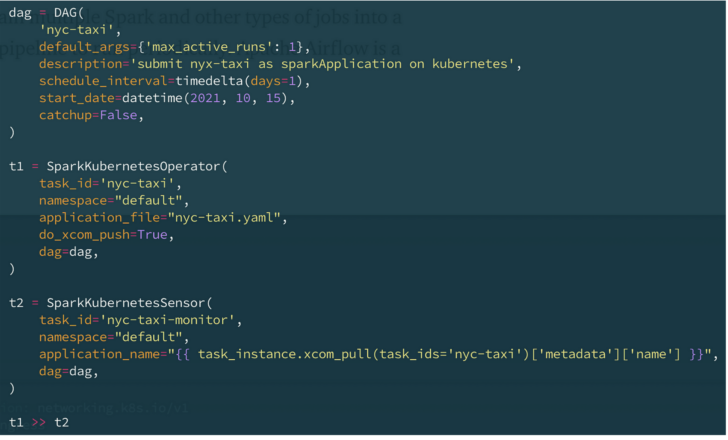
A simple pipeline defined in Airflow.
On the Airflow UI, this is what the DAG looks like:
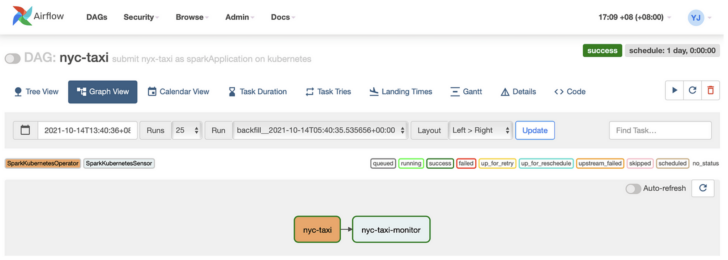
The NYC Taxi analysis DAG on Airflow UI.
While running, the first task in the DAG will spin up multiple Apache Spark pods, as defined in the nyc-taxi.yaml file, on Kubernetes through the Spark on k8s operator, just like the kubectl apply command does.
Spark pods spun up by Airflow.
Airflow helps manage dependencies and scheduling of the multi-job workflow. Since it reuses the jobs and runs in the same Kubernetes environment, overhead of introducing Airflow is minimal.
Conclusion
With Apache Spark on Kubernetes, and FlashBlade S3 object storage for the data, my data engineering process is dramatically simplified. I would open my browser to start quick prototyping and data exploration. Thanks to the power of Spark on Kubernetes, I don’t have to limit my prototyping and exploration to a small set of sample data. Once I am good with the prototype, I put the code in a Python file, modify, and submit it for running in production with a single Kubernetes command. For complex pipelines, I would orchestrate the jobs with Airflow to help manage dependencies and scheduling.
This is my data engineering workflow. I like that I only need a single environment for all of these.
Pure Storage Analytics and AI Solutions
![]()